Tutorial¶
Exploring EGADS¶
The simplest way to start working with EGADS is to run it from the Python command line. To load EGADS into the Python name-space, simply import it:
>>> import egads
You may then begin working with any of the algorithms and functions contained in EGADS.
There are several useful methods to explore the routines contained in EGADS.
The first is using the Python built-in dir() command:
>>> dir(egads)
returns all the classes and subpackages contained in EGADS. EGADS follows the naming conventions from the Python Style Guide (http://www.python.org/dev/peps/pep-0008), so classes are always MixedCase, functions and modules are generally lowercase or lowercase_with_underscores. As a further example,
>>> dir(egads.input)
would returns all the classes and subpackages of the egads.input module.
Another way to explore EGADS is by using tab completion, if supported by your Python installation. Typing
>>> egads.
then hitting TAB will return a list of all available options.
Python has built-in methods to display documentation on any function known as docstrings.
The easiest way to access them is using the help() function:
>>> help(egads.input.NetCdf)
or
>>> egads.input.NetCdf?
will return all methods and their associated documentation for the NetCdf class.
Simple operations with EGADS¶
To have a list of file in a directory, use the following function:
>>> egads.input.get_file_list('path/to/all/netcdf/files/*.nc')
The EgadsData class¶
At the core of the EGADS package is a data class intended to handle data and associated metadata in a consistent way between files, algorithms and within the framework. This ensures that important metadata is not lost when combining data form various sources in EGADS.
Additionally, by subclassing the Quantities and Numpy packages, EgadsData incorporates unit comprehension to reduce unit-conversion errors during calculation, and supports broad array manipulation capabilities. This section describes how to employ the EgadsData class in the EGADS program scope.
Creating EgadsData instances¶
The EgadsData class takes four basic arguments:
- value
- Value to assign to
EgadsDatainstance. Can be scalar, array, or otherEgadsDatainstance.
- units
- Units to assign to
EgadsDatainstance. Should be string representation of units, and can be a compound units type such as ‘g/kg’, ‘m/s^2’, ‘feet/second’, etc.
- variable metadata
- An instance of the
VariableMetadatatype or dictionary, containing keywords and values of any metadata to be associated with thisEgadsDatainstance.
- other attributes
- Any other attributes added to the class are automatically stored in the
VariableMetadatainstance associated with theEgadsDatainstance.
The following are examples of creating EgadsData instances:
>>> x = egads.EgadsData([1,2,3], 'm')
>>> a = [1,2,3,4]
>>> b = egads.EgadsData(a, 'km', b_metadata)
>>> c = egads.EgadsData(28, 'degC', long_name="current temperature")
If, during the call to EgadsData, no units are provided, but a variable metadata instance is provided with a units property, this will then be used to define the EgadsData units:
>>> x_metadata = egads.core.metadata.VariableMetadata({'units':'m', 'long_name':'Test Variable'})
>>> x = egads.EgadsData([1,2,3], x_metadata)
>>> print(x.units)
m
>>> print(x.metadata)
{'units': 'm', 'long_name': 'Test Variable'}
The EgadsData is a subclass of the Quantities and Numpy packages. Thus it allows different kind of operations like addition, substraction, slicing, and many more. For each of those operations, a new EgadsData class is created with all their attributes.
Note
With mathematical operands, as metadata define an EgadsData before an operation, and may not reflect the new EgadsData, it has been decided to not keep the metadata attribute. It’s the responsability of the user to add a new VariableMetadata instance or a dictionary to the new EgadsData object. It is not true if a user wants to slice an EgadsData. In that case, metadata are automatically attributed to the new EgadsData.
Metadata¶
The metadata object used by EgadsData is an instance of VariableMetadata, a dictionary object containing methods to recognize, convert and validate known metadata types. It can reference parent metadata objects, such as those from an algorithm or data file, to enable users to track the source of a particular variable.
When reading in data from a supported file type (NetCDF, NASA Ames, Hdf), or doing calculations with an EGADS algorithm, EGADS will automatically populate the associated metadata and assign it to the output variable. However, when creating an EgadsData instance manually, the metadata must be user-defined.
As mentioned, VariableMetadata is a dictionary object, thus all metadata are stored as keyword:value pairs. To create metadata manually, simply pass in a dictionary object containing the desired metadata:
>>> var_metadata_dict = {'long_name':'test metadata object', '_FillValue':-9999}
>>> var_metadata = egads.core.metadata.VariableMetadata(var_metadata_dict)
To take advantage of its metadata recognition capabilities, a conventions keyword can be passed with the variable metadata to give a context to these metadata.
>>> var_metadata = egads.core.metadata.VariableMetadata(var_metadata_dict, conventions='CF-1.0')
If a particular VariableMetadata object comes from a file or algorithm, the class attempts to assign the conventions automatically. While reading from a file, for example, the class attempts to discover the conventions used based on the “Conventions” keyword, if present.
Working with units¶
EgadsData subclasses Quantities, thus all of the latter’s unit comprehension methods are available when using EgadsData. This section will outline the basics of unit comprehension. A more detailed tutorial of the unit comprehension capabilities can be found at https://python-quantities.readthedocs.io/en/latest
In general, units are assigned to EgadsData instances when they are being created.
>>> a = egads.EgadsData([1,2,3], 'm')
>>> b = egads.EgadsData([4,5,6], 'meters/second')
Once a unit type has been assigned to an EgadsData instance, it will remain that class of unit and can only be converted between other types of that same unit. The rescale method can be used to convert between similar units, but will give an error if an attempt is made to convert to non-compatible units.
>>> a = egads.EgadsData([1,2,3], 'm')
>>> a_km = a.rescale('km')
>>> print(a_km)
['EgadsData', array([0.001, 0.002, 0.003]), 'km']
>>> a_grams = a.rescale('g')
ValueError: Unable to convert between units of "m" and "g"
Likewise, arithmetic operations between EgadsData instances are handled using the unit comprehension provided by Quantities. For example adding units of a similar type is permitted:
>>> a = egads.EgadsData(10, 'm')
>>> b = egads.EgadsData(5, 'km')
>>> a + b
['EgadsData', array(5010.0), 'm']
But, non-compatible types cannot be added. They can, however, be multiplied or divided:
>>> distance = egads.EgadsData(10, 'm')
>>> time = egads.EgadsData(1, 's')
>>> distance + time
ValueError: Unable to convert between units of "s" and "m"
>>> distance/time
['EgadsData', array(10), 'm/s']
Working with raw text files¶
EGADS provides the egads.input.text_file_io.EgadsFile class as a simple wrapper for interacting with generic text files. EgadsFile can read, write and display data from text files, but does not have any tools for automatically formatting input or output data.
Opening¶
To open a text file, simply create a EgadsFile instance with the parameters filename and perms:
>>> import egads
>>> f = egads.input.EgadsFile('/pathname/filename.nc', 'r')
-
EgadsFile(filename[, perms='r'])¶ Open a text file.
Parameters: - filename (string) – path and filename of a text file
- perms (string) – permissions ; optional
Return type: text file
Valid values for permissions are:
r– Read: opens file for reading only. Default value if nothing is provided.w– Write: opens file for writing, and overwrites data in file.a– Append: opens file for appending data.r+– Read and write: opens file for both reading and writing.
File Manipulation¶
The following methods are available to control the current position in the file and display more information about the file.
-
f.display_file()¶ Prints contents of the file out to a standard output.
-
f.get_position()¶ Returns the current position in the file as an integer.
-
f.seek(location[, from_where='b'])¶ Seeks to a specified location in the text file.
Parameters: - location (int) – it is an integer specifying how far to seek
- from_where (string) – it is an option to specify from where to seek, valid options for from_where are
bto seek from beginning of file,cto seek from current position in file andeto seek from the end of the file ; optional
Return type: position in the text file
-
f.reset()¶ Resets the position to the beginning of the file.
Reading Data¶
Reading data is done using the read(size) method on a file that has been opened with r or r+ permissions:
>>> import egads
>>> f = egads.input.EgadsFile()
>>> f.open('myfile.txt','r')
>>> single_char_value = f.read()
>>> multiple_chars = f.read(10)
If the size parameter is not specified, the read() function will input a single character from the open file. Providing an integer value n as the size parameter to read(size) will return n characters from the open file.
Data can be read line-by-line from text files using read_line():
>>> line_in = f.read_line()
Writing Data¶
To write data to a file, use the write(data) method on a file that has been opened with w, a or r+ permissions:
>>> import egads
>>> f = egads.input.EgadsFile()
>>> f.open('myfile.txt','a')
>>> data = 'Testing output data to a file.\n This text will appear on the 2nd line.'
>>> f.write(data)
Tutorial¶
Here is a basic ASCII file, created by EGADS:
# The current file has been created with EGADS
# Institution: My Institution
# Author(s): John Doe
time sea level corr sea level
1.0 5.0 1.0
2.0 2.0 3.0
3.0 -2.0 -1.0
4.0 0.5 2.5
5.0 4.0 6.0
This file has been created with the following commands:
import EGADS module:
>>> import egads
create two main variables, following the official EGADS convention:
>>> data1 = egads.EgadsData(value=[5.0,2.0,-2.0,0.5,4.0], units='mm', standard_name='sea level', scale_factor=1., add_offset=0., _FillValue=-9999) >>> data2 = egads.EgadsData(value=[1.0,3.0,-1.0,2.5,6.0], units='mm', standard_name='corr sea level', scale_factor=1., add_offset=0., _FillValue=-9999)
create an independant variable, still by following the official EGADS convention:
>>> time = egads.EgadsData(value=[1.0,2.0,3.0,4.0,5.0], units='seconds since 19700101T00:00:00', standard_name='time')
create a new EgadsFile instance:
>>> f = egads.input.EgadsFile()
use the following function to open a new file:
>>> f.open('main_raw_file.dat', 'w')
prepare the headers if necessary:
>>> headers = '# The current file has been created with EGADS\n# Institution: My Institution\n# Author(s): John Doe\n' >>> headers += time.metadata["standard_name"] + ' ' + data1.metadata["standard_name"] + ' ' + data2.metadata["standard_name"])
prepare an object to receive all data:
>>> data = '' >>> for i, _ in enumerate(time.value): ... data += str(time.value[i]) + ' ' + str(data1.value[i]) + ' ' + str(data2.value[i]) + '\n'
write the headers and data into the file
>>> f.write(headers) >>> f.write(data)
and do not forget to close the file:
>>> f.close()
Working with CSV files¶
egads.input.text_file_io.EgadsCsv is designed to easily input or output data in CSV format. Data input using EgadsCsv is separated into a list of arrays, which each column a separate array in the list.
Opening¶
To open a csv file, simply create a EgadsCsv instance with the parameters filename, perms, delimiter and quotechar:
>>> import egads
>>> f = egads.input.EgadsCsv('/pathname/filename.nc', 'r', ',','"')
-
EgadsCsv(filename[, perms='r', delimiter=', ', quotechar='"'])¶ Open a text file.
Parameters: - filename (string) – path and filename of a text file
- perms (string) – permissions ; optional
- delimiter (string) – a one-character string used to separate fields ; optional
- quotechar (string) – a one-character string used to quote fields containing special characters ; optional
Return type: csv file
Valid values for permissions are:
r– Read: opens file for reading only. Default value if nothing is provided.w– Write: opens file for writing, and overwrites data in file.a– Append: opens file for appending data.r+– Read and write: opens file for both reading and writing.
File Manipulation¶
The following methods are available to control the current position in the file and display more information about the file.
-
f.display_file() Prints contents of the file out to a standard output.
-
f.get_position() Returns the current position in the file as an integer.
-
f.seek(location[, from_where='b']) Seeks to a specified location in the text file.
Parameters: - location (int) – it is an integer specifying how far to seek
- from_where (string) – it is an option to specify from where to seek, valid options for from_where are
bto seek from beginning of file,cto seek from current position in file andeto seek from the end of the file ; optional
Return type: position in the text file
-
f.reset() Resets the position to the beginning of the file.
Reading Data¶
Reading data is done using the read(lines, format) method on a file that has been opened with r or r+ permissions:
>>> import egads
>>> f = egads.input.EgadsCsv()
>>> f.open('mycsvfile.csv','r')
>>> single_line_as_list = f.read(1)
>>> all_lines_as_list = f.read()
-
f.read([lines=None, format=None])¶ Returns a list of items read in from the CSV file.
Parameters: - lines (int) – if it is provided, the function will read in the specified number of lines, otherwise it will read the whole file ; optional
- format (string) – it is an optional list of characters used to decompose the elements read in from the CSV files to their proper types, options are ; optional
Return type: list of items read in from the CSV file
Valid options for format:
i– intf– floatl– longs– string
Thus to read in the line:
FGBTM,20050105T143523,1.5,21,25
the command to input with proper formatting would look like this:
>>> data = f.read(1, ['s','s','f','f'])
Writing Data¶
To write data to a file, use the write(data) method on a file that has been opened with w, a or r+ permissions:
>>> import egads
>>> f = egads.input.EgadsCsv()
>>> f.open('mycsvfile.csv','a')
>>> titles = ['Aircraft ID','Timestamp','Value1','Value2','Value3']
>>> f.write(titles)
where the titles parameter is a list of strings. This list will be output to the CSV, with each strings separated by the delimiter specified when the file was opened (default is ,).
To write multiple lines out to a file, writerows(data) is used:
>>> data = [['FGBTM','20050105T143523',1.5,21,25],['FGBTM','20050105T143524',1.6,20,25.6]]
>>> f.writerows(data)
Tutorial¶
Here is a basic CSV file, created by EGADS:
time,sea level,corrected sea level
1.0,5.0,1.0
2.0,2.0,3.0
3.0,-2.0,-1.0
4.0,0.5,2.5
5.0,4.0,6.0
This file has been created with the following commands:
import EGADS module:
>>> import egads
create two main variables, following the official EGADS convention:
>>> data1 = egads.EgadsData(value=[5.0,2.0,-2.0,0.5,4.0], units='mm', standard_name='sea level', scale_factor=1., add_offset=0., _FillValue=-9999) >>> data2 = egads.EgadsData(value=[1.0,3.0,-1.0,2.5,6.0], units='mm', standard_name='corr sea level', scale_factor=1., add_offset=0., _FillValue=-9999)
create an independant variable, still by following the official EGADS convention:
>>> time = egads.EgadsData(value=[1.0,2.0,3.0,4.0,5.0], units='seconds since 19700101T00:00:00', standard_name='time')
create a new EgadsFile instance:
>>> f = egads.input.EgadsCsv()
use the following function to open a new file:
>>> f.open('main_csv_file.csv','w',',','"')
prepare the headers if necessary:
>>> headers = ['time', 'sea level', 'corrected sea level']
prepare an object to receive all data:
>>> data = [time.value, data1.value, data2.value]
write the headers and data into the file
>>> f.write(headers) >>> f.write(data)
and do not forget to close the file:
>>> f.close()
Working with NetCDF files¶
EGADS provides two classes to work with NetCDF files. The simplest, egads.input.netcdf_io.NetCdf, allows simple read/write operations to NetCDF files. The other, egads.input.netcdf_io.EgadsNetCdf, is designed to interface with NetCDF files conforming to the EUFAR Standards & Protocols data and metadata regulations. This class directly reads or writes NetCDF data using instances of the EgadsData class.
Opening¶
To open a NetCDF file, simply create a EgadsNetCdf instance or a NetCdf instance with the parameters filename and perms:
>>> import egads
>>> f = egads.input.EgadsNetCdf('/pathname/filename.nc', 'r')
-
EgadsNetCdf(filename[, perms='r'])¶ Open a NetCDF file conforming the the EUFAR Standards & Protocols data and metadata regulations.
Parameters: - filename (string) – path and filename of a NetCDF file
- perms (string) – permissions ; optional
Return type: NetCDF file.
Valid values for permissions are:
r– Read: opens file for reading only. Default value if nothing is provided.w– Write: opens file for writing, and overwrites data in file.a– Append: opens file for appending data.r+– Same asa.
Getting info¶
-
f.get_dimension_list([varname=None, group_walk=False, details=False])¶ Returns a dictionary of all dimensions with their sizes. varname is optional and can take three different forms: varname is a variable name, and in that case the function returns a dictionary of all dimensions and their sizes attached to varname at the root of the NetCDF file ; varname is a path to a group + a variable name, the function returns a dictionary of all dimensions and their sizes attached to varname in the specified group ; varname is a path to group + a group name, the function returns a dictionary of all dimensions and their sizes in the specified group. group_walk is optional and if True, the function will explore the entire file is varname is None, or from a specified group if varname is a group path. details is optional, and if True, the path of each dimension is included in the final dictionary.
Parameters: - varname (string) – Name of variable or group to get the list of associated dimensions from. If no variable name is provided, the function returns all dimensions at the root of the NetCDF file ; optional
- group_walk (bool) – if True, the function visits all groups (if at least one exists) to list all dimensions. False by default ; optional
- details (bool) – if True, dimension path is provided in the dictionary. False by default ; optional
Return type: ordered dictionary of dimensions
>>> print(f.get_dimension_list('temperature')) >>> print(f.get_dimension_list('test1/data/temperature', details=True)) >>> print(f.get_dimension_list('test1/data', group_walk=True))
-
f.get_attribute_list([varname=None])¶ Returns a list of all top-level attributes. varname is optional and can take three different forms: varname is a variable name, and in that case the function returns all attributes and their values attached to varname at the root of the NetCDF file ; varname is a path to a group + a variable name, the function returns all attributes and their values attached to varname in the specified group ; varname is a path to group + a group name, the function returns all attributes and their values attached to the specified group.
Parameters: varname (string) – Name of variable or group to get the list of attributes from. If no variable name is provided, the function returns top-level NetCDF attributes ; optional Return type: dictionary of attributes >>> print(f.get_attribute_list('temperature')) >>> print(f.get_attribute_list('test1/data/temperature')) >>> print(f.get_attribute_list('test1/data'))
-
f.get_variable_list([groupname=None, group_walk=False, details=False])¶ Returns a list of all variables at the root of the NetCDF file. If groupname is provided, the function will list all variables located at groupname. If group_walk is True, the function will list all variables in the NetCDF file from root, or from groupname if groupname is provided, to the last folder. If details is True, the function returns a list of dictionary containing the name of the variable and its path in the NetCDF file. By default, details is False and the function returns a simple list of variable names.
Parameters: - groupname (string) – the name of the group to get the variable list from ; optional
- group_walk (bool) – if True, the function lists all variables from the root of the file, or from groupname if provided, to the last group ; optional
- details (bool) – if True, the function returns a list of dictionary in which the key is the name of the variable, and the value is the path of the variable in the NetCDF file ; optional
Return type: list of variables
>>> print(f.get_variable_list()) >>> print(f.get_variable_list('test1/data')) >>> print(f.get_variable_list('test1/data', group_walk=True, details=True))
-
f.get_group_list([groupname=None, details=False])¶ Returns a list of groups found in the NetCDF file. If groupname is provided, the function returns all groups from groupname to the last group in groupname. The function returns a list of string if details is False. If details is True, it returns a list of dictionary in which the key is the name of the group and the value its path in the NetCDF file.
Parameters: - groupname (string) – name of a group where to get the group list ; optional
- details (bool) – if True, the function returns a list of dictionary in which the key is the name of the group and the value its path in the NetCDF file ; optional
Return type: list of strings or list of dictionary
>>> print(f.get_group_list()) >>> print(f.get_group_list('test1', True))
-
f.get_filename()¶ Returns the filename for the currently opened file.
Return type: filename
-
f.get_perms()¶ Returns the current permissions on the file that is open.
Return type: permissions
Reading data¶
To read data from a file, use the read_variable() function:
>>> data = f.read_variable(varname, input_range, read_as_float, replace_fill_value)
-
f.read_variable(varname[, input_range=None, read_as_float=False, replace_fill_value=False])¶ If using the
NetCdfclass, an array of values contained in varname will be returned. If using theEgadsNetCdfclass, an instance of theEgadsDataclass will be returned containing the values and attributes of varname. If a group path is present in varname, then the function reads the variable varname in that specified group.Parameters: - varname (string) – name of a variable, with or without group path, in the NetCDF file
- input_range (list) – list of min/max values ; optional
- read_as_float (bool) – if True, EGADS reads the data and convert them to float numbers, if False, the data type is the type of data in file ; optional
- replace_fill_value (bool) – if True, EGADS reads the data and replace
_FillValueormissing_value(if one of the attributes exists) in data by NaN (numpy.nan) ; optional
Return type: data,
EgadsDataor array>>> data = f.read_variable('temperature') >>> data = f.read_variable('test1/data/temperature')
Writing data¶
The following describe how to add dimensions or attributes to a file.
-
f.add_dim(name, size)¶ - Add a dimension to the NetCDF file. If a path to a group plus the dimension name is included in name, the dimension is
- added to the group. In that case, the group has to be created before.
Parameters: - name (string) – the name of the dimension
- size (int) – the size of the dimension
>>> f.add_dim('time', len(time)) >>> f.add_dim('time', len(time), 'test1/data') >>> f.add_dim('time', len(time), ['test1', 'data'])
-
f.add_attribute(attrname, value[, varname=None])¶ Add an attribute to the NetCDF file. If varname is None, the attribute is a global attribute, and if not, the attribute is a variable attribute attached to varname. If the path of a group is present in varname, the attribute is attached to the variable stored in the specified group. If varname is simply the path of a group, attribute is attached to the group.
Parameters: - attrname (string) – the name of the attribute
- value (string|float|int) – the value of the attribute
- varname (string) – the name of the variable | group to which to attach the attribute ; optional
>>> f.add_attribute('project', 'my project') >>> f.add_attribute('project', 'my project', 'temperature') >>> f.add_attribute('project', 'my project', 'test1/data/temperature') >>> f.add_attribute('project', 'my project', 'test1/data')
If using EgadsNetCdf, data can be output to variables using the write_variable() function as follows:
>>> f.write_variable(data, varname, dims, ftype, fillvalue)
-
f.write_variable(data, varname[, dims=None, ftype='double', fillvalue=None])¶ Write the values contained in data in the variable varname in a NetCDF file. If varname contains a path to a group, the variable will be created in the specified group, but in that case the group has to be created before. An instance of
EgadsDatamust be passed intowrite_variable. Any attributes that are contained within theEgadsDatainstance are applied to the NetCDF variable as well. If an attribute with a name equal to_FillValueormissing_valueis found, NaN in data will be automatically replaced by the missing value. If attributes with the namescale_factorand/oradd_offsetare found, those attributes are automatically applied to the data.Parameters: - data (EgadsData|array|vector|scalar) – values to be stored in the NetCDF file
- varname (string) – the name of the variable, or the path of group + the name of the variable, in the NetCDF file
- dims (tuple) – a tuple of dimension names for data (not needed if the variable already exists) ; optional
- ftype (string) – the data type of the variable, the default value is double, other valid options are float, int, short, char and byte ; optional
- fillvalue (float|int) – if it is provided, it overrides the default NetCDF _FillValue ; optional, it doesn’t exist if using
EgadsNetCdf
>>> f.write_variable(data, 'particle_size', ('time', )) >>> f.write_variable(data, 'test1/data/particle_size', ('time', ))
If using NetCdf, data can be output to variables using the write_variable() function as follows:
>>> f.write_variable(data, varname, dims, ftype, fillvalue, scale_factor, add_offset)
-
f.write_variable(data, varname[, dims=None, ftype='double', fillvalue=None, scale_factor=None, add_offset=None]) Write the values contained in data in the variable varname in a NetCDF file. If varname contains a path to a group, the variable will be created in the specified group, but in that case the group has to be created before. Values for data passed into
write_variablemust be scalar or array.Parameters: - data (EgadsData|array|vector|scalar) – values to be stored in the NetCDF file
- varname (string) – the name of the variable, or the path of group + the name of the variable, in the NetCDF file
- dims (tuple) – a tuple of dimension names for data (not needed if the variable already exists) ; optional
- ftype (string) – the data type of the variable, the default value is double, other valid options are float, int, short, char and byte ; optional
- fillvalue (float|int) – if it is provided, it overrides the default NetCDF _FillValue ; optional, it doesn’t exist if using
EgadsNetCdf - scale_factor (float|int) – if data must be scaled, use this parameter ; optional
- add_offset (float|int) – if an offset must be added to data, use this parameter ; optional
>>> f.write_variable(data, 'particle_size', ('time', )) >>> f.write_variable(data, 'test1/data/particle_size', ('time', ), scale_factor=1.256, add_offset=5.56)
Conversion from NetCDF to NASA Ames file format¶
The conversion is only possible on opened NetCDF files and with variables at the root of the NetCDF file. If modifications have been made and haven’t been saved, the conversion won’t take into account those modifications. Actually, the only File Format Index supported by the conversion is 1001. Consequently, if more than one independant variables are present in the NetCDF file, the file won’t be converted and the function will raise an exception. If the user needs to convert a complex file with variables depending on multiple independant variables, and with the presence of groups, the conversion should be done manually by creating a NasaAmes instance and a NasaAmes dictionary, by populating the dictionary and by saving the file.
To convert a NetCDF file to NasaAmes file format, simply use:
-
f.convert_to_nasa_ames([na_file=None, float_format=None, delimiter=' ', no_header=False])¶ Convert the opened NetCDF file to NasaAmes file.
Parameters: - na_file (string) – it is the name of the output file once it has been converted, by default, na_file is None, and the name of the NetCDF file will be used with the extension .na ; optional
- float_format (string) – it is the formatting string used for formatting floats when writing to output file ; optional
- delimiter (string) – it is a character or a sequence of character to use between data items in the data file ; optional (by default ‘ ‘, 4 spaces)
- no_header (bool) – if it is set to
True, then only the data blocks are written to file ; optional
>>> f.convert_to_nasa_ames(na_file='nc_converted_to_na.na', float_format='%.8f', delimiter=';', no_header=False)
To convert a NetCDF file to NasaAmes CSV file format, simply use:
-
f.convert_to_csv([csv_file=None, float_format=None, no_header=False])¶ Convert the opened NetCDF file to NasaAmes CSV file.
Parameters: - csv_file (string) – it is the name of the output file once it has been converted, by default, na_file is None, and the name of the NetCDF file will be used with the extension .csv ; optional
- float_format (string) – it is the formatting string used for formatting floats when writing to output file ; optional
- no_header (bool) – if it is set to
True, then only the data blocks are written to file ; optional
Conversion from NetCDF to Hdf5 file format¶
EGADS Lineage offers a direct possibility to convert a full NetCDF file to Hdf file format. In the case of complexe NetCdf files, a manual Hdf file creation and editing is still possible.
-
f.convert_to_hdf([filename=None])¶ Converts the opened NetCdf file to Hdf format following the EUFAR and EGADS convention. If groups exist, they are preserved in the new Hdf file.
Parameters: filename (string) – if only a name is given, a Hdf file named filenameis created in the NetCdf file folder ; if a path and a name are given, a Hdf file namednameis created in the folderpath; optional
Other operations¶
-
f.get_attribute_value(attrname[, varname=None])¶ Return the value of the global attribute attrname, or the value of the variable attribute attrname if varname is not None. If varname contains a path to a group + a variable name, the function returns the attribute value attached to the variable in the specified group. If varname is simple path of group, the functions returns the attribute value attached to the group.
Parameters: - attrname (string) – the name of the attribute
- varname (string) – the name of the variable | group to which the attribute is attached
Return type: value of the attribute
>>> print(f.get_attribute_value('project')) >>> print(f.get_attribute_value('long_name', 'temperature')) >>> print(f.get_attribute_value('long_name', 'test1/data/temperature')) >>> print(f.get_attribute_value('project', 'test1/data'))
-
f.change_variable_name(varname, newname)¶ Change a variable name in the currently opened NetCDF file. If varname contains a path to a group + a variable name, the variable name in the specified group is changed.
Parameters: - attrname (string) – the actual name of the variable
- varname (string) – the new name of the variable
>>> f.change_variable_name('particle_nbr', 'particle_number') >>> f.change_variable_name('test1/data/particle_nbr', 'particle_number')
-
f.add_group(groupname)¶ Create a group in the NetCDF file. groupname can be a path + a group name or a sequence of group, in both cases, intermediary groups are created if needed.
Parameters: groupname (string|list) – a group name or a list of group name >>> f.add_group('MSL/north_atlantic/data') >>> f.add_group(['MSL', 'north_atlantic', 'data'])
Closing¶
To close a file, simply use the close() method:
>>> f.close()
Note
The EGADS NetCdf and EgadsNetCdf use the official NetCDF I/O routines, therefore, as described in the NetCDF documentation, it is not possible to remove a variable or more, and to modify the values of a variable. As attributes, global and those linked to a variable, are more dynamic, it is possible to remove, rename, or replace them.
Tutorial¶
Here is a NetCDF file, created by EGADS, and viewed by the command ncdump -h ....:
=> ncdump -h main_netcdf_file.nc
netcdf main_netcdf_file {
dimensions:
time = 5 ;
variables:
double time(time) ;
time:units = "seconds since 19700101T00:00:00" ;
time:long_name = "time" ;
double sea_level(time) ;
sea_level:_FillValue = -9999. ;
sea_level:category = "TEST" ;
sea_level:scale_factor = 1. ;
sea_level:add_offset = 0. ;
sea_level:long_name = "sea level" ;
sea_level:units = "mm" ;
double corrected_sea_level(time) ;
corrected_sea_level:_FillValue = -9999. ;
corrected_sea_level:units = "mm" ;
corrected_sea_level:add_offset = 0. ;
corrected_sea_level:scale_factor = 1. ;
corrected_sea_level:long_name = "corr sea level" ;
// global attributes:
:Conventions = "CF-1.0" ;
:history = "the netcdf file has been created by EGADS" ;
:comments = "no comments on the netcdf file" ;
:institution = "My institution" ;
}
This file has been created with the following commands:
import EGADS module:
>>> import egads
create two main variables, following the official EGADS convention:
>>> data1 = egads.EgadsData(value=[5.0,2.0,-2.0,0.5,4.0], units='mm', standard_name='sea level', scale_factor=1., add_offset=0., _FillValue=-9999) >>> data2 = egads.EgadsData(value=[1.0,3.0,-1.0,2.5,6.0], units='mm', standard_name='corr sea level', scale_factor=1., add_offset=0., _FillValue=-9999)
create an independant variable, still by following the official EGADS convention:
>>> time = egads.EgadsData(value=[1.0,2.0,3.0,4.0,5.0], units='seconds since 19700101T00:00:00', standard_name='time')
create a new EgadsNetCdf instance with a file name:
>>> f = egads.input.EgadsNetCdf('main_netcdf_file.nc', 'w')
add the global attributes to the NetCDF file:
>>> f.add_attribute('Conventions', 'CF-1.0') >>> f.add_attribute('history', 'the netcdf file has been created by EGADS') >>> f.add_attribute('comments', 'no comments on the netcdf file') >>> f.add_attribute('institution', 'My institution')
add the dimension(s) of your variable(s), here it is
time:>>> f.add_dim('time', len(time))
write the variable(s), it is a good practice to write at the first place the independant variable
time:>>> f.write_variable(time, 'time', ('time',), 'double') >>> f.write_variable(data1, 'sea_level', ('time',), 'double') >>> f.write_variable(data2, 'corrected_sea_level', ('time',), 'double')
and do not forget to close the file:
>>> f.close()
Working with Hdf files¶
EGADS provides two classes to work with Hdf files. The simplest, egads.input.hdf_io.Hdf, allows simple read/write operations to Hdf files. The other, egads.input.hdf_io.EgadsHdf, is designed to interface with Hdf files conforming to the EUFAR Standards & Protocols data and metadata regulations. This class directly reads or writes Hdf data using instances of the EgadsData class.
Opening¶
To open a Hdf file, simply create a EgadsHdf instance or a Hdf instance with the parameters filename and perms:
>>> import egads
>>> f = egads.input.EgadsHdf('/pathname/filename.nc', 'r')
-
EgadsHdf(filename[, perms='r'])¶ Open a Hdf file conforming the the EUFAR Standards & Protocols data and metadata regulations.
Parameters: - filename (string) – path and filename of a Hdf file
- perms (string) – permissions ; optional
Return type: Hdf file.
Valid values for permissions are:
r– Read: opens file for reading only. Default value if nothing is provided.w– Write: opens file for writing, and overwrites data in file.a– Append: opens file for appending data.r+– Same asa.
Getting info¶
-
f.get_dimension_list([varname=None, group_walk=False, details=False]) Returns a dictionary of all dimensions with their sizes. varname is optional and can take three different forms: varname is a variable name, and in that case the function returns a dictionary of all dimensions and their sizes attached to varname at the root of the Hdf file ; varname is a path to a group + a variable name, the function returns a dictionary of all dimensions and their sizes attached to varname in the specified group ; varname is a path to a group + a group name, the function returns a dictionary of all dimensions and their sizes in the specified group. group_walk is optional and if True, the function will explore the entire file is varname is None, or from a specified group if varname is a group path. details is optional, and if True, the path of each dimension is included in the final dictionary.
Parameters: - varname (string) – Name of variable or group to get the list of associated dimensions from. If no variable name is provided, the function returns all dimensions at the root of the Hdf file ; optional
- group_walk (bool) – if True, the function visits all groups (if at least one exists) to list all dimensions. False by default ; optional
- details (bool) – if True, dimension path is provided in the dictionary. False by default ; optional
Return type: ordered dictionary of dimensions
>>> print(f.get_dimension_list('temperature')) >>> print(f.get_dimension_list('test1/data/temperature')) >>> print(f.get_dimension_list('test1/data'))
-
f.get_attribute_list([objectname=None]) Returns a list of all top-level attributes. varname is optional and can take three different forms: objectname is a variable name, and in that case the function returns all attributes and their values attached to objectname at the root of the Hdf file ; objectname is a path to a group + a variable name, the function returns all attributes and their values attached to objectname in the specified group ; objectname is a path to a group + a group name, the function returns all attributes and their values attached to the specified group.
Parameters: objectname (string) – name of a variable / group ; optional Return type: dictionary of attributes >>> print(f.get_attribute_list('temperature')) >>> print(f.get_attribute_list('test1/data/temperature')) >>> print(f.get_attribute_list('test1/data'))
-
f.get_variable_list([groupname=None, group_walk=False, details=False]) Returns a list of all variables at the root of the Hdf file if groupname is None, otherwise a list of all variables in the group groupname. If group_walk is True, the the function will explore all the file or from groupname if groupname is provided. If details is True, the function returns a list of dictionary containing the name of the variable and its path in the Hdf file. By default, details is False and the function returns a simple list of variable names.
Parameters: - groupname (string) – the name of the group to get the list from ; optional
- group_walk (bool) – if True, the function visits all groups (if at least one exists) to list all variables. False by default ; optional
- details (bool) – if True, the function returns a list of dictionary in which the key is the name of the variable, and the value is the path of the variable in the Hdf file ; optional
Return type: list of variables
>>> print(f.get_variable_list()) >>> print(f.get_variable_list(details=True))
-
f.get_file_structure()¶ Returns a view of the file structure, groups and datasets.
Return type: list of strings and Hdf objects >>> print(f.get_file_structure())
-
f.get_group_list([groupname=None, details=False]) Returns a list of groups found in the Hdf file. If groupname is provided, the function returns all groups from groupname to the last group in groupname. The function returns a list of string if details is False. If details is True, it returns a list of dictionary in which the key is the name of the group and the value its path in the Hdf file.
Parameters: - groupname (string) – name of a group where to get the group list ; optional
- details (bool) – if True, the function returns a list of dictionary in which the key is the name of the group and the value its path in the Hdf file ; optional
Return type: list of strings or list of dictionary
>>> print(f.get_group_list()) >>> print(f.get_group_list('test1', True))
-
f.get_filename() Returns the filename for the currently opened file.
Return type: filename
-
f.get_perms() Returns the current permissions on the file that is open.
Return type: permissions
Reading data¶
To read data from a file, use the read_variable() function:
>>> data = f.read_variable(varname, input_range, read_as_float, replace_fill_value)
-
f.read_variable(varname[, input_range=None, read_as_float=False, replace_fill_value=False]) If using the
Hdfclass, an array of values contained in varname will be returned. If using theEgadsHdfclass, an instance of theEgadsDataclass will be returned containing the values and attributes of varname. If a group path is present in varname, then the function reads the variable varname in that specified group.Parameters: - varname (string) – name of a variable, with or without group path, in the Hdf file
- input_range (list) – list of min/max values ; optional
- read_as_float (bool) – if True, EGADS reads the data and convert them to float numbers, if False, the data type is the type of data in file ; optional
- replace_fill_value (bool) – if True, EGADS reads the data and replace
_FillValueormissing_value(if one of the attributes exists) in data by NaN (numpy.nan) ; optional
Return type: data,
EgadsDataor array>>> data = f.read_variable('temperature') >>> data = f.read_variable('test1/data/temperature')
Writing data¶
The following describe how to add dimensions or attributes to a file.
-
f.add_dim(name, data[, ftype='double']) Add a dimension to the Hdf file. The name of the dimension can include a path to a group where to store the dimension. In that case, the group has to be created before. If using
Hdf, values for data passed intoadd_dimmust be scalar or array. Otherwise, if usingEgadsHdf, an instance ofEgadsDatamust be passed intoadd_dim. In this case, any attributes that are contained within theEgadsDatainstance are applied to the Hdf variable as well.Parameters: - name (string) – the name of the dimension, or the path to a group + the name of the dimension
- data (EgadsData|array|vector|scalar) – values to be stored in the Hdf file
- ftype (string) – the data type of the variable, the default value is double, other valid options are float, int, short, char and byte ; optional
>>> f.add_dim('time', time_data, len(time)) >>> f.add_dim('test1/data/time', time_data, len(time))
-
f.add_attribute(attrname, value[, objname=None]) Add an attribute to the Hdf file. If objname is None, the attribute is a global attribute, and if not, the attribute is attached to objname. objname can be a group (with or without a path) or variable (with or without a path).
Parameters: - attrname (string) – the name of the attribute
- value (string|float|int) – the value of the attribute
- objname (string) – the name of the variable | group to which to attach the attribute ; optional
>>> f.add_attribute('project', 'my project') >>> f.add_attribute('project', 'my project', 'temperature') >>> f.add_attribute('project', 'my project', 'test1/data/temperature') >>> f.add_attribute('project', 'my project', 'test1/data')
Data can be output to variables using the write_variable() function as follows:
>>> f.write_variable(data, varname, dims, ftype, fillvalue)
-
f.write_variable(data, varname[, dims=None, ftype='double']) Write the values contained in data in the variable varname in a Hdf file. If varname contains a path to a group, the variable will be created in the specified group, but in that case the group has to be created before. If using
Hdf, values for data passed intowrite_variablemust be scalar or array. Otherwise, if usingEgadsHdf, an instance ofEgadsDatamust be passed intowrite_variable. In this case, any attributes that are contained within theEgadsDatainstance are applied to the Hdf variable as well.Parameters: - data (EgadsData|array|vector|scalar) – values to be stored in the Hdf file
- varname (string) – the name of the variable, or the path of group + the name of the variable, in the Hdf file
- dims (tuple) – a tuple of dimension names for data (not needed if the variable already exists) ; optional
- ftype (string) – the data type of the variable, the default value is double, other valid options are float, int, short, char and byte ; optional
>>> f.write_variable(data, 'particle_size', ('time', )) >>> f.write_variable(data, 'test1/data/particle_size', ('time', ))
Conversion from Hdf5 to NASA Ames file format¶
The conversion is only possible on opened Hdf files and with variables at the root of the Hdf file. If modifications have been made and haven’t been saved, the conversion won’t take into account those modifications. Actually, the only File Format Index supported by the conversion is 1001. Consequently, if more than one independant variables are present in the Hdf file, the file won’t be converted and the function will raise an exception. If the user needs to convert a complex file with variables depending on multiple independant variables, and with the presence of groups, the conversion should be done manually by creating a NasaAmes instance and a NasaAmes dictionary, by populating the dictionary and by saving the file.
To convert a Hdf file to NasaAmes file format, simply use:
-
f.convert_to_nasa_ames([na_file=None, float_format=None, delimiter=' ', no_header=False]) Convert the opened NetCDF file to NasaAmes file.
Parameters: - na_file (string) – it is the name of the output file once it has been converted, by default, na_file is None, and the name of the Hdf file will be used with the extension .na ; optional
- float_format (string) – it is the formatting string used for formatting floats when writing to output file ; optional
- delimiter (string) – it is a character or a sequence of character to use between data items in the data file ; optional (by default ‘ ‘, 4 spaces)
- no_header (bool) – if it is set to
True, then only the data blocks are written to file ; optional
>>> f.convert_to_nasa_ames(na_file='nc_converted_to_na.na', float_format='%.8f', delimiter=';', no_header=False)
To convert a Hdf file to NasaAmes CSV file format, simply use:
-
f.convert_to_csv([csv_file=None, float_format=None, no_header=False]) Convert the opened Hdf file to NasaAmes CSV file.
Parameters: - csv_file (string) – it is the name of the output file once it has been converted, by default, na_file is None, and the name of the Hdf file will be used with the extension .csv ; optional
- float_format (string) – it is the formatting string used for formatting floats when writing to output file ; optional
- no_header (bool) – if it is set to
True, then only the data blocks are written to file ; optional
Conversion from Hdf5 to NetCDF file format¶
EGADS Lineage offers a direct possibility to convert a full Hdf file to NetCDF file format. In the case of complexe Hdf5 files, a manual NetCDF file creation and editing is still possible.
-
f.convert_to_netcdf([filename=None])¶ Converts the opened Hdf file to NetCdf format following the EUFAR and EGADS convention. If groups exist, they are preserved in the new NetCDF file.
Parameters: filename (string) – if only a name is given, a NetCDF file named filenameis created in the HDF file folder ; if a path and a name are given, a NetCDF file namednameis created in the folderpath; optional
Other operations¶
-
f.get_attribute_value(attrname[, objectname=None]) Return the value of the global attribute attrname, or the value of the variable attribute attrname if objectname is not None. If objectname contains a path to a group + a variable name, the function returns the attribute value attached to the variable in the specified group. If objectname is simple path of group, the functions returns the attribute value attached to the group.
Parameters: - attrname (string) – the name of the attribute
- objectname (string) – the name of the variable | group to which the attribute is attached
Return type: value of the attribute
>>> print(f.get_attribute_value('project')) >>> print(f.get_attribute_value('long_name', 'temperature')) >>> print(f.get_attribute_value('long_name', 'test1/data/temperature')) >>> print(f.get_attribute_value('project', 'test1/data'))
-
f.add_group(groupname) Create a group in the Hdf file. groupname can be a path + a group name or a sequence of group, in both cases, intermediary groups are created if needed.
Parameters: groupname (string|list) – a group name or a list of group name >>> f.add_group('MSL/north_atlantic/data') >>> f.add_group(['MSL', 'north_atlantic', 'data'])
-
f.delete_attribute(attrname[, objectname=None])¶ Delete the attribute attrname at the root of the Hdf file if objectname is None, or attached to objectname. objectname can be the name of a variable or a group, or a path to a group plus the name of a variable or a group.
Parameters: - attrname (string) – the name of the attribute
- objectname (string) – the name of the variable | group to which the attribute is attached
>>> f.delete_attribute('long_name') >>> f.delete_attribute('long_name', 'temperature') >>> f.delete_attribute('long_name', 'test1/data/temperature') >>> f.delete_attribute('project', 'test1/data')
-
f.delete_group(groupname)¶ Delete the group groupname in the Hdf file. groupname can be a name of a group at the root of the Hdf file, or a path to a group plus the name of a a group.
Parameters: attrname (string) – the name of the group >>> f.delete_group('data') >>> f.delete_group('test1/data')
-
f.delete_variable(varname)¶ Delete the variable varname in the Hdf file. varname can be the name of a variable or a path to a group plus the name of a variable.
Parameters: varname (string) – the name of the variable >>> f.delete_variable('temperature') >>> f.delete_variable('test1/data/temperature')
Dataset with compound data¶
Dataset with compound data are not specifically handled by EGADS. When a dataset is read and contains compound data, it is possible to access the different fields in this way:
>>> temperature = f.read_variable('temperature')
>>> date_field = temperature['date']
Obviously, if the user declared an EgadsHdf instance to read the file, the compound data is still an EgadsData instance, with the same metadata and units. If the user declared an Hdf instance to read the file, the compound data is a Numpy ndarray instance. In EGADS, units are handled automatically. Thus, with compound data and multiple data in the same dataset, it is highly likely that units won’t be handled properly, and will be set to dimensionless most of the time.
Note
With the instance EgadsHdf, an attribute has been added to the EgadsData instance, compound_data, which informs the user about the dataset type. If the dataset contains compound data, the attribute compound_data is set to True ; if not it is set to False. Then it is the responsability of the user to explore the different fields in the dataset.
Tutorial¶
Here is a Hdf file, created by EGADS, and viewed by the command ncdump -h ....:
=> ncdump -h main_hdf_file.hdf5
netcdf main_hdf_file {
dimensions:
time = 5 ;
variables:
double corrected_sea_level(time) ;
string corrected_sea_level:name = "corr sea level" ;
corrected_sea_level:scale_factor = 1. ;
corrected_sea_level:_FillValue = -9999 ;
string corrected_sea_level:units = "mm" ;
double sea_level(time) ;
string sea_level:name = "sea level" ;
sea_level:scale_factor = 1. ;
sea_level:_FillValue = -9999 ;
string sea_level:units = "mm" ;
double time(time) ;
string time:name = "time" ;
string time:units = "seconds since 19700101T00:00:00" ;
// global attributes:
string :Conventions = "CF-1.0" ;
string :history = "the hdf file has been created by EGADS" ;
string :comments = "no comments on the hdf file" ;
string :institution = "My institution" ;
}
This file has been created with the following commands:
import EGADS module:
>>> import egads
create two main variables, following the official EGADS convention:
>>> data1 = egads.EgadsData(value=[5.0,2.0,-2.0,0.5,4.0], units='mm', name='sea level', scale_factor=1., add_offset=0., _FillValue=-9999) >>> data2 = egads.EgadsData(value=[1.0,3.0,-1.0,2.5,6.0], units='mm', name='corr sea level', scale_factor=1., add_offset=0., _FillValue=-9999)
create an independant variable, still by following the official EGADS convention:
>>> time = egads.EgadsData(value=[1.0,2.0,3.0,4.0,5.0], units='seconds since 19700101T00:00:00', name='time')
create a new EgadsHdf instance with a file name:
>>> f = egads.input.EgadsHdf('main_hdf_file.hdf5', 'w')
add the global attributes to the Hdf file:
>>> f.add_attribute('Conventions', 'CF-1.0') >>> f.add_attribute('history', 'the hdf file has been created by EGADS') >>> f.add_attribute('comments', 'no comments on the hdf file') >>> f.add_attribute('institution', 'My institution')
add the dimension(s) of your variable(s), here it is
time:>>> f.add_dim('time', time)
write the variable(s), and no need to write the variable
time, it has already been added by the commandadd_dim():>>> f.write_variable(data1, 'sea_level', ('time',), 'double') >>> f.write_variable(data2, 'corrected_sea_level', ('time',), 'double')
and do not forget to close the file:
>>> f.close()
Working with NASA Ames files¶
EGADS provides two classes to work with NASA Ames files. The simplest, egads.input.nasa_ames_io.NasaAmes, allows simple read/write operations. The other, egads.input.nasa_ames_io.EgadsNasaAmes, is designed to interface with NASA Ames files conforming to the EUFAR Standards & Protocols data and metadata regulations. This class directly reads or writes NASA Ames file using instances of the EgadsData class. Actually, only the FFI 1001 has been interfaced with EGADS.
Opening¶
To open a NASA Ames file, simply create a EgadsNasaAmes instance with the parameters pathname and permissions:
>>> import egads
>>> f = egads.input.EgadsNasaAmes('/pathname/filename.na','r')
-
EgadsNasaAmes(pathname[, permissions='r'])¶ Open a NASA Ames file conforming the the EUFAR Standards & Protocols data and metadata regulations.
Parameters: - filename (string) – path and filename of a NASA Ames file
- perms (string) – permissions ; optional
Return type: NasaAmes file.
Valid values for permissions are:
r– Read: opens file for reading only. Default value if nothing is provided.w– Write: opens file for writing, and overwrites data in file.a– Append: opens file for appending data.r+– Same asa.
Once a file has been opened, a dictionary of NASA/Ames format elements is loaded into memory. That dictionary will be used to overwrite the file or to save a new file.
Getting info¶
-
f.get_dimension_list([na_dict=None]) Returns a list of all variable dimensions.
Parameters: na_dict (dict) – if provided, the function get dimensions from the NasaAmes dictionary na_dict, if not dimensions are from the opened file ; optional Return type: dictionary of dimensions
-
f.get_attribute_list([varname=None, vartype='main', na_dict=None]) Returns a dictionary of all top-level attributes.
Parameters: - varname (string) – name of a variable, if provided, the function returns a dictionary of all attributes attached to varname ; optional
- vartype (string) – if provided and varname is not
None, the function will search in the variable type vartype by default ; optional - na_dict (dict) – if provided, it will return a list of all top-level attributes, or all varname attributes, from the NasaAmes dictionary na_dict ; optional
Return type: dictionary of attributes
-
f.get_attribute_value(attrname[, varname=None, vartype='main', na_dict=None]) Returns the value of a top-level attribute named attrname.
Parameters: - attrname (string) – the name of the attribute
- varname (string) – name of a variable, if provided, the function returns the value of the attribute attached to varname ; optional
- vartype (string) – if provided and varname is not
None, the function will search in the variable type vartype by default ; optional - na_dict (dict) – if provided, it will return the value of an attribute from the NasaAmes dictionary na_dict ; optional
Return type: value of attribute
-
f.get_variable_list([na_dict=None]) Returns a list of all variables.
Parameters: na_dict (dict) – if provided, it will return the list of all variables from the NasaAmes dictionary na_dict ; optional Return type: list of variables
-
f.get_filename() Returns the filename for the currently opened file.
Return type: filename
-
f.get_perms() Returns the current permissions on the file that is open.
Return type: permissions
Reading data¶
To read data from a file, use the read_variable() function:
>>> data = f.read_variable(varname, na_dict, read_as_float, replace_fill_value)
-
f.read_variable(varname[, na_dict=None, read_as_float=False, replace_fill_value=False]) If using the
NasaAmesclass, an array of values contained in varname will be returned. If using theEgadsNasaAmesclass, an instance of theEgadsDataclass will be returned containing the values and attributes of varname.Parameters: - varname (string) – name of a variable in the NasaAmes file
- na_dict (dict) – it will tell to EGADS in which Nasa Ames dictionary to read data, if na_dict is
None, data are read in the opened file ; optional - read_as_float (bool) – if True, EGADS reads the data and convert them to float numbers, if False, the data type is the type of data in file ; optional
- replace_fill_value (bool) – if True, EGADS reads the data and replace
_FillValueormissing_value(if one of the attributes exists) in data by NaN (numpy.nan) ; optional
Return type: data,
EgadsDataor array
Writing data¶
To write data to the current file or to a new file, the user must save a dictionary of NasaAmes elements. Few functions are available to help him to prepare the dictionary:
-
f.create_na_dict()¶ Create a new dictionary populated with standard NasaAmes keys
-
f.write_attribute_value(attrname, attrvalue[, na_dict=None, varname=None, vartype='main'])¶ Write or replace a specific attribute (from the official NasaAmes attribute list) in the currently opened dictionary.
Parameters: - attrname (string) – name of the attribute in the NasaAmes dictionary
- attrvalue (string|float|integer|list|array) – value of the attribute
- na_dict (dict) – if provided the function will write the attribute in the NasaAmes dictionary na_dict ; optional
- varname (string) – if provided, write or replace a specific attribute linked to the variable var_name in the currently opened dictionary ; accepted attributes for a variable are ‘name’, ‘units’, ‘_FillValue’ and ‘scale_factor’, other attributes will be refused and should be passed as ‘special comments’ ; optional
- vartype (string) – if provided and varname is not
None, the function will search in the variable type vartype by default ; optional
-
f.write_variable(data[, varname=None, vartype='main', attrdict=None, na_dict=None]) Write or replace a variable in the currently opened dictionary. If using the
NasaAmesclass, an array of values for data is asked. If using theEgadsNasaAmesclass, an instance of theEgadsDataclass must be injected for data. If aEgadsDatais passed into thewrite_variablefunction, any attributes that are contained within theEgadsDatainstance are automatically populated in the NASA Ames dictionary as well, those which are not mandatory are stored in the ‘SCOM’ attribute. If an attribute with a name equal to_FillValueormissing_valueis found, NaN in data will be automatically replaced by the missing value.Parameters: - data (EgadsData|array|vector|scalar) – values to be stored in the NasaAmes file
- varname (string) – the name of the variable ; if data is an
EgadsData, mandatory if ‘standard_name’ or ‘long_name’ is not an attribute of data ; absolutely mandatory if data is not anEgadsData; optional - vartype (string) – the type of data, ‘independant’ or ‘main’, only mandatory if data must be stored as an independant variable (dimension) ; optional
- attrdict (dict) – a dictionary containing mandatory attributes (‘name’, ‘units’, ‘_FillValue’ and ‘scale_factor’), only mandatory if data is not an
EgadsData; optional - na_dict (dict) – if provided, the function stores the variable in the NasaAmes dictionary na_dict
Saving a file¶
Once a dictionary is ready, use the save_na_file() function to save the file:
>>> data = f.save_na_file(filename, na_dict, float_format, delimiter, no_header):
-
f.save_na_file([filename=None, na_dict=None, float_format=None, delimiter=' ', no_header=False])¶ Save the opened NasaAmes dictionary and file.
Parameters: - filename (string) – is the name of the new file, if not provided, the name of the opened NasaAmes file is used ; optional
- na_dict (dict) – the name of the NasaAmes dictionary to be saved, if not provided, the opened dictionary will be used ; optional
- float_format (string) – the format of the floating numbers in the file (by default, no round up) ; optional
- delimiter (string) – it is a character or a sequence of character to use between data items in the data file ; optional (by default ‘ ‘, 4 spaces)
- no_header (bool) – if it is set to
True, then only the data blocks are written to file ; optional
Conversion from NASA/Ames file format to NetCDF¶
When a NASA/Ames file is opened, all metadata and data are read and stored in memory in a dedicated dictionary. The conversion will convert that dictionary to generate a NetCDF file. If modifications are made to the dictionary, the conversion will take into account those modifications. Actually, the only File Format Index supported by the conversion in the NASA Ames format is 1001. Consequently, if variables depend on multiple independant variables (e.g. data is function of time, longitude and latitude), the file won’t be converted and the function will raise an exception. If the user needs to convert a complex file with variables depending on multiple independant variables, the conversion should be done manually by creating a NetCDF instance and by populating the NetCDF files with NASA/Ames data and metadata.
To convert a NASA/Ames file, simply use:
-
f.convert_to_netcdf([nc_file=None, na_dict=None]) Convert the opened NasaAmes file to NetCDF file format.
Parameters: - nc_file (string) – if provided, the function will use nc_file for the path and name of the new_file, if not, the function will take the name and path of the opened NasaAmes file and replace the extension by ‘.nc’ ; optional
- na_dict (dict) – the name of the NasaAmes dictionary to be converted, if not provided, the opened dictionary will be used ; optional
Conversion from NASA/Ames file format to Hdf5¶
When a NASA/Ames file is opened, all metadata and data are read and stored in memory in a dedicated dictionary. The conversion will convert that dictionary to generate a Hdf file. If modifications are made to the dictionary, the conversion will take into account those modifications. Actually, the only File Format Index supported by the conversion in the NASA Ames format is 1001. Consequently, if variables depend on multiple independant variables (e.g. data is function of time, longitude and latitude), the file won’t be converted and the function will raise an exception. If the user needs to convert a complex file with variables depending on multiple independant variables, the conversion should be done manually by creating a Hdf instance and by populating the Hdf files with NASA/Ames data and metadata.
To convert a NASA/Ames file, simply use:
-
f.convert_to_hdf([hdf_file=None, na_dict=None]) Convert the opened NasaAmes file to Hdf file format.
Parameters: - hdf_file (string) – if provided, the function will use hdf_file for the path and name of the new_file, if not, the function will take the name and path of the opened NasaAmes file and replace the extension by ‘.h5’ ; optional
- na_dict (dict) – the name of the NasaAmes dictionary to be converted, if not provided, the opened dictionary will be used ; optional
Other operations¶
-
f.read_na_dict()¶ Returns a deep copy of the current opened file dictionary
Return type: deep copy of a dictionary
-
egads.input.nasa_ames_io.na_format_information()¶ Returns a text explaining the structure of a NASA/Ames file to help the user to modify or to create his own dictionary
Return type: string
Tutorial¶
Here is a NASA/Ames file:
23 1001
John Doe
An institution
tide gauge
ATESTPROJECT
1 1
2017 1 30 2017 1 30
0.0
time (seconds since 19700101T00:00:00)
2
1 1
-9999 -9999
sea level (mm)
corr sea level (mm)
3
========SPECIAL COMMENTS===========
this file has been created with egads
=========END=========
4
========NORMAL COMMENTS===========
headers:
time sea level corrected sea level
=========END=========
1.00 5.00 1.00
2.00 2.00 3.00
3.00 -2.00 -1.00
4.00 0.50 2.50
5.00 4.00 6.00
This file has been created with the following commands:
import EGADS module:
>>> import egads
create two main variables, following the official EGADS convention:
>>> data1 = egads.EgadsData(value=[5.0,2.0,-2.0,0.5,4.0], units='mm', name='sea level', scale_factor=1, _FillValue=-9999) >>> data2 = egads.EgadsData(value=[1.0,3.0,-1.0,2.5,6.0], units='mm', name='corr sea level', scale_factor=1, _FillValue=-9999)
create an independant variable, still by following the official EGADS convention:
>>> time = egads.EgadsData(value=[1.0,2.0,3.0,4.0,5.0], units='seconds since 19700101T00:00:00', name='time')
create a new NASA/Ames empty instance:
>>> f = egads.input.NasaAmes()
initialize a new NASA/Ames dictionary:
>>> na_dict = f.create_na_dict()
prepare the normal and special comments if needed, in a list, one cell for each line, or only one string with lines separated by
\n:>>> scom = ['========SPECIAL COMMENTS===========','this file has been created with egads','=========END========='] >>> ncom = ['========NORMAL COMMENTS===========','headers:','time sea level corrected sea level','=========END========='] or >>> scom = '========SPECIAL COMMENTS===========\nthis file has been created with egads\n=========END=========' >>> ncom = '========NORMAL COMMENTS===========\nheaders:\ntime sea level corrected sea level\n=========END========='
populate the main NASA/Ames attributes:
>>> f.write_attribute_value('ONAME', 'John Doe', na_dict = na_dict) # ONAME is the name of the author(s) >>> f.write_attribute_value('ORG', 'An institution', na_dict = na_dict) # ORG is tne name of the organization responsible for the data >>> f.write_attribute_value('SNAME', 'tide gauge', na_dict = na_dict) # SNAME is the source of data (instrument, observation, platform, ...) >>> f.write_attribute_value('MNAME', 'ATESTPROJECT', na_dict = na_dict) # MNAME is the name of the mission, campaign, programme, project dedicated to data >>> f.write_attribute_value('DATE', [2017, 1, 30], na_dict = na_dict) # DATE is the date at which the data recorded in this file begin (YYYY MM DD) >>> f.write_attribute_value('NIV', 1, na_dict = na_dict) # NIV is the number of independent variables >>> f.write_attribute_value('NSCOML', 3, na_dict = na_dict) # NSCOML is the number of special comments lines or the number of elements in the SCOM list >>> f.write_attribute_value('NNCOML', 4, na_dict = na_dict) # NNCOML is the number of special comments lines or the number of elements in the NCOM list >>> f.write_attribute_value('SCOM', scom, na_dict = na_dict) # SCOM is the special comments attribute >>> f.write_attribute_value('NCOM', ncom, na_dict = na_dict) # NCOM is the normal comments attribute
write each variable in the dictionary:
>>> f.write_variable(time, 'time', vartype="independant", na_dict = na_dict) >>> f.write_variable(data1, 'sea level', vartype="main", na_dict = na_dict) >>> f.write_variable(data2, 'corrected sea level', vartype="main", na_dict = na_dict)
and finally, save the dictionary to a NASA/Ames file:
>>> f.save_na_file('na_example_file.na', na_dict)
Working with algorithms¶
Algorithms in EGADS are stored in the egads.algorithms module for embedded algorithms and in egads.user_algorithms module for user-defined algorithms. They are separated into sub-modules by category (microphysics, thermodynamics, radiation, etc). Each algorithm follows a standard naming scheme, using the algorithm’s purpose and source:
{CalculatedParameter}{Detail}{Source}
For example, an algorithm which calculates static temperature, which was provided by CNRM would be named:
TempStaticCnrm
Getting algorithm information¶
There are several methods to get information about each algorithm contained in EGADS. The EGADS Algorithm Handbook is available for easy reference outside of Python. In the handbook, each algorithm is described in detail, including a brief algorithm summary, descriptions of algorithm inputs and outputs, the formula used in the algorithm, algorithm source and links to additional references. The handbook also specifies the exact name of the algorithm as defined in EGADS. The handbook can be found on the EGADS website.
Within Python, usage information on each algorithm can be found using the help() command:
>>> help(egads.algorithms.thermodynamics.VelocityTasCnrm)
>>> Help on class VelocityTasCnrm in module egads.algorithms.thermodynamics.
velocity_tas_cnrm:
class VelocityTasCnrm(egads.core.egads_core.EgadsAlgorithm)
| FILE velocity_tas_cnrm.py
|
| VERSION Revision: 1.02
|
| CATEGORY Thermodynamics
|
| PURPOSE Calculate true airspeed
|
| DESCRIPTION Calculates true airspeed based on static temperature,
| static pressure and dynamic pressure using St Venant's
| formula.
|
| INPUT T_s vector K or C static temperature
| P_s vector hPa static pressure
| dP vector hPa dynamic pressure
| cpa coeff. J K-1 kg-1 specific heat of air (dry
| air is 1004 J K-1 kg-1)
| Racpa coeff. () R_a/c_pa
|
| OUTPUT V_p vector m s-1 true airspeed
|
| SOURCE CNRM/GMEI/TRAMM
|
| REFERENCES "Mecanique des fluides", by S. Candel, Dunod.
|
| Bulletin NCAR/RAF Nr 23, Feb 87, by D. Lenschow and
| P. Spyers-Duran
|
...
Calling algorithms¶
Algorithms in EGADS generally accept and return arguments of EgadsData type, unless otherwise noted. This has the advantages of constant typing between algorithms, and allows metadata to be passed along the whole processing chain. Units on parameters being passed in are also checked for consistency, reducing errors in calculations, and rescaled if needed. However, algorithms will accept any normal data type, as well. They can also return non-EgadsData instances, if desired.
To call an algorithm, simply pass in the required arguments, in the order they are described in the algorithm help function. An algorithm call, using the VelocityTasCnrm in the previous section as an example, would therefore be the following:
>>> V_p = egads.algorithms.thermodynamics.VelocityTasCnrm().run(T_s, P_s, dP,
cpa, Racpa)
where the arguments T_s, P_s, dP, etc are all assumed to be previously defined in the program scope. In this instance, the algorithm returns an EgadsData instance to V_p. To run the algorithm, but return a standard data type (scalar or array of doubles), set the return_Egads flag to false.
>>> V_p = egads.algorithms.thermodynamics.VelocityTasCnrm(return_Egads=False).
run(T_s, P_s, dP, cpa, Racpa)
If an algorithm has been created by a user and is not embedded by default in EGADS, it should be called like this:
>>> V_p = egads.user_algorithms.thermodynamics.VelocityTasCnrm().run(T_s, P_s, dP,
cpa, Racpa)
Note
When injecting a variable in an EgadsAlgorithm, the format of the variable should follow closely the documentation of the algorithm. If the variable is a scalar, and the algorithm needs a vector, the scalar should be surrounded by brackets: 52.123 -> [52.123].
Scripting¶
The recommended method for using EGADS is to create script files, which are extremely useful for common or repetitive tasks. This can be done using a text editor of your choice. The example script belows shows the calculation of density for all NetCDF files in a directory.
#!/usr/bin/env python
# import egads package
import egads
# import thermodynamic module and rename to simplify usage
import egads.algorithms.thermodynamics as thermo
# get list of all NetCDF files in 'data' directory
filenames = egads.input.get_file_list('data/*.nc')
f = egads.input.EgadsNetCdf() # create EgadsNetCdf instance
for name in filenames: # loop through files
f.open(name, 'a') # open NetCdf file with append permissions
T_s = f.read_variable('T_t') # read in static temperature
P_s = f.read_variable('P_s') # read in static pressure from file
rho = thermo.DensityDryAirCnrm().run(P_s, T_s) # calculate density
f.write_variable(rho, 'rho', ('Time',)) # output variable
f.close() # close file
Scripting Hints¶
When scripting in Python, there are several important differences from other programming languages to keep in mind. This section outlines a few of these differences.
Importance of white space¶
Python differs from C++ and Fortran in how loops or nested statements are signified. Whereas C++ uses brackets (’{’ and ‘}’) and FORTRAN uses end statements to signify the end of a nesting, Python uses white space. Thus, for statements to nest properly, they must be set at the proper depth. As long as the document is consistent, the number of spaces used doesn’t matter, however, most conventions call for 4 spaces to be used per level. See below for examples:
FORTRAN:
X = 0
DO I = 1,10
X = X + I
PRINT I
END DO
PRINT X
Python:
x = 0
for i in range(1,10):
x += i
print i
print x
Using the GUI¶
Since September 2016, a Graphical User Interface is available at https://github.com/eufarn7sp/egads-gui. It gives the user the possibility to explore data, apply/create algorithms, display and plot data.
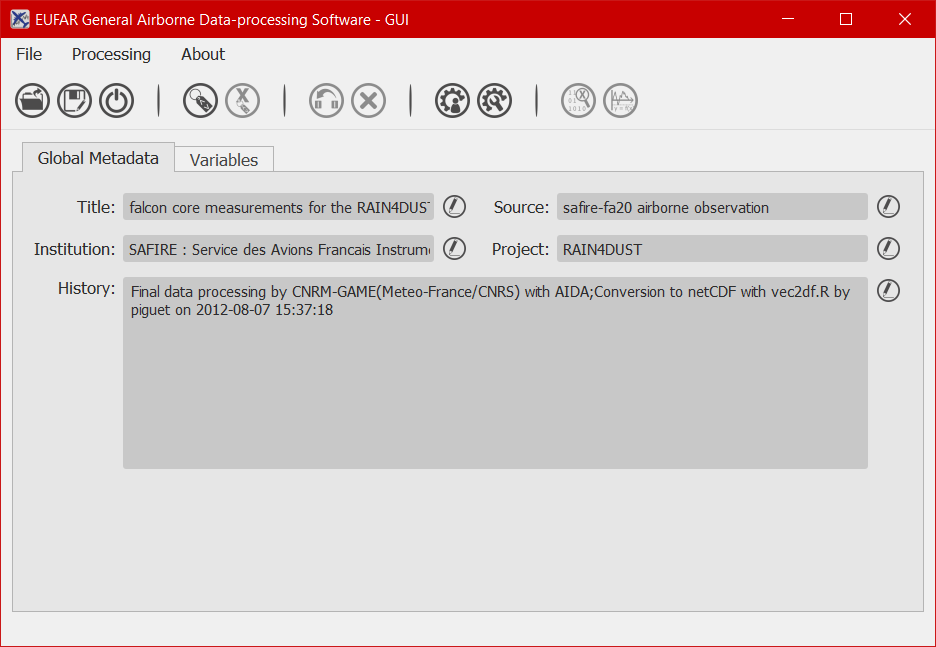
EGADS GUI can be launched as a simple python script from the terminal, if EGADS is installed, and once in the EGADS GUI directory:
>>> python egads_gui.py
Since version 1.0.0, a stand-alone package is available for those who wants to use the GUI without a Python installation. In that case, look for EGADS Lineage GUI STA in the release part of the repository. For Windows (from Windows 7 32), donwload the .msi package and launch the installation, it should be installed outside ProgramFiles to avoid issues with admin rights, then the GUI can be run by double clicking on egads_gui.exe or from the shortcut in the Startup menu. A .zip package is also available for those who don’t want to install it. For Linux (from Linux 4.15), download the tar.gz package somewhere on your hard drive (preferably in your home directory), extract it and run egads_gui.
The stand-alone versions for Linux and Windows have been created with PyInstaller, Windows 7 and Ubuntu 18.04.
Note
As for EGADS, the Graphical User Interface is available from two branches: master and Lineage (https://github.com/EUFAR/egads-gui/tree/Lineage). The Lineage one is only compatible with Python 3 and the earlier versions of EGADS Lineage.